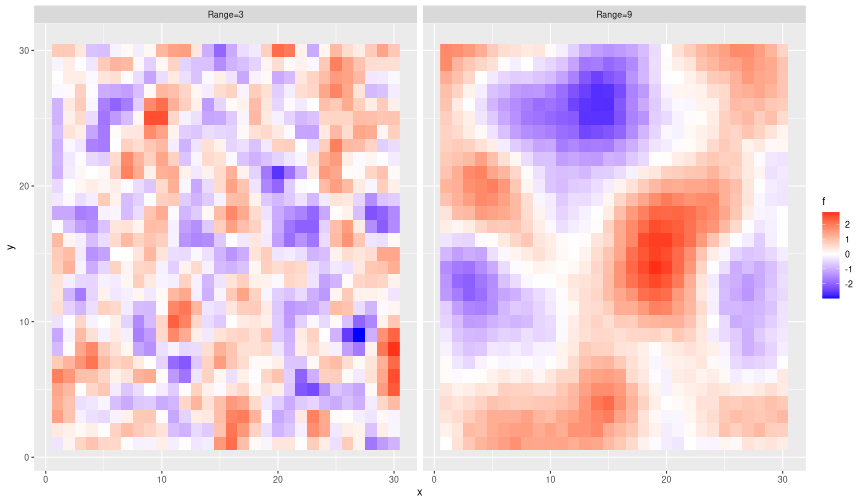
set.seed(123) n = 30 # define grid coordinates xy_grid = crossing(x=1:n, y=1:n) # use `dist` to calculate matrix of euclidean distances d = as.matrix(dist(xy_grid)) # exponential covariance function k = function(range=1, sill=1, nugget=0.01) { return(sill * exp(-3*(d/range)^2) + diag(nugget, nrow(d))) } # draw two realisations with different range parameters gp_sim = xy_grid %>% mutate(`Range=3` = drop(rmvnorm(n=1, sigma=k(range=3))), `Range=9` = drop(rmvnorm(n=1, sigma=k(range=9)))) %>% pivot_longer(c(-x,-y), names_to='range', values_to='f') # visualise them on a grid ggplot(gp_sim) + geom_raster(aes(x=x, y=y, fill=f)) + facet_wrap(~range) + scale_fill_gradient2(low='blue', high='red')