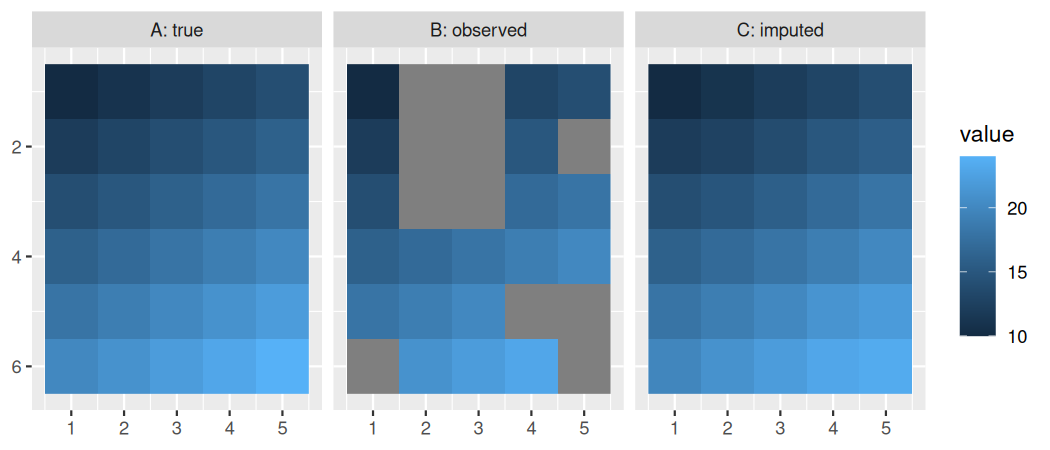
# plot results crossing(jj=1:nx, ii=1:ny) %>% mutate(`A: true` = c(x), `B: observed` = c(y), `C: imputed` = c(y_imputed)) %>% pivot_longer(-c(jj,ii), names_to='type', values_to='value') %>% ggplot() + geom_raster(aes(x=jj, y=ii, fill=value)) + facet_wrap(~type) + scale_y_reverse() + labs(x=NULL, y=NULL)